DNA replication is a complex process of synthesis of DNA molecules. This process occurs in the S phase of the cell cycle. Each DNA molecule forms two new daughter DNA molecules.
Three possible patterns of the DNA replication process are
proposed. The actual DNA replication mode is called the semiconservative model.
The two others are theoretical.
a.
Semiconservative DNA replication
According to this model, the parental DNA molecule helix unwinds and
unzips, and both the strands acting as a template synthesize the new daughter
DNA. The new strands are synthesized complementary to both old strands. If T
(thymidylic acid) is present, A (adenylic acid) is added; if G (guanidylic
acid) is present, C (cytidylic acid) is added; likewise, A would attract T, and
C would attract G. Each replicated DNA molecule would consist of one “old” and
one “new” strand, hence the reason for the name semiconservative
replication. Watson and Crick
proposed this model for DNA replication. They proposed that DNA replication is
semiconservative.
b.
Conservative DNA replication
This theoretical mode of replication
also relies on the parental strands as a template. According to this model, the complementary polynucleotide chains
are synthesized. Following synthesis, however, the two newly created strands
then come together, and the parental strands also recombine. The original DNA
helix is thus “conserved.”
c.
Dispersive DNA replication
According to this theoretical mode, the replication of new DNA also
relies on the parental strands as a template. In this model, the parental
strands are dispersed into two new double helices following replication. Hence,
each strand consists of both old and new DNA. This mode would involve cleavage
of the parental strands during replication.
Meselson-Stahl experiment- Replication as a semi-conservative process
Matthew Meselson and Franklin Stahl in 1958, provided strong evidence that semiconservative replication is the actual mode used by cells to produce new DNA molecules. They experimented on E. coli cells in a medium that had 15NH4Cl (ammonium chloride) as a source of heavy nitrogen containing one more neutron than the naturally occurring 14N isotope.
Mathew Meselson and Franklin grew bacteria with 14N in their DNA in a growth medium with a heavy isotope of 15N nitrogen for many generations. After many generations, almost all the bacteria had heavier isotope 15N in their DNA. The newly formed bacteria were shifted to a 14N medium containing only 14NH4Cl. These cell samples are then removed back from the medium and centrifuged. Three separate samples were obtained. One sample was obtained just after the cells were introduced into the 14N medium. This sample was named 0 sample. The second sample was obtained just after 20 minutes, called sample 20. The other sample was obtained after another 20 minutes, called sample 40 minutes.
DNA samples were obtained from all the bacteria and were dissolved into cesium chloride and then spun at a very high speed in an ultra-centrifuge for many hours. The DNA of normal bacteria appeared the lightest as formed sediment at the top of the test tube, while the DNA of sample 0 minute appeared heaviest as it formed sediment at the bottom of the test tube. The DNA sample at 20 minutes formed sediment intermediate level to that of the natural sample and 0-minute sample. The sample 40 minutes had two sediments, one at the top and the other at the intermediate level.
Meselson and Stahl interpreted their results as follows; The DNA of the control sample had both the strands of 14N, whereas the DNA sample of 0 minutes had both the strands 15N and the DNA of the sample 20 minutes had one strand of 14N, and the other of 15N.
Process of DNA replication
The process of replication involves the following main steps
a. Uncoiling of DNA helix
DNA has a helical structure. Before
replication, Topoisomerase, also called DNA gyrase, makes a single-strand
cut and hydrolyzes the phosphodiester linkage. It causes the uncoiling of the
double helix.
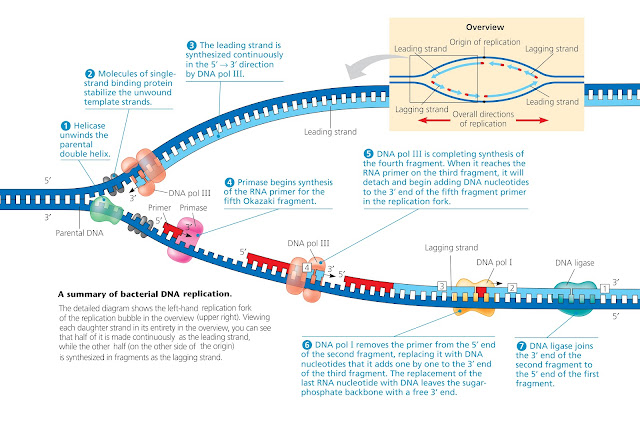
b. Unwinding of the duplex
After uncoiling helicase breaks the
hydrogen bond between the nitrogen bases forming the replication bubble. The
ends of the replication bubble are called replication fork.
Both strands tend to reunite forming
a duplex. Therefore, to prevent the formation of the duplex SSB proteins
bind to 8-10 nucleotides on a single strand of the two strands.
d. Assembly of primer
The key enzyme of the
replication process is the DNA polymerase III. This enzyme cannot add the 1st
nucleotide and requires a preexisting nucleotide in the new DNA strand. This is
provided by a primase enzyme which synthesizes an RNA sequence of about 10-30
basses RNA primer. The primer is a small segment that provides the OH end for
the next nucleotide.
The DNA polymerase-III adds complementary nucleotides to both the template strands. The DNA polymerase remains in the replication fork on the template strand and continuously adds nucleotides in 5-3 directions to the new strand as the fork progresses. The two strands of a double helix are antiparallel to each other, one runs in the 5′ to 3′ direction, while the other has the opposite 3′ to 5′ polarity. DNA Pol III synthesizes DNA in only the 5′ to 3′ direction, therefore the direction of synthesis on both strands is opposite to each other.
One strand
is synthesized towards the replication fork is called the leading strand having
continuous DNA. The other strand that is synthesized in the opposite direction
to the replication fork is called the lagging strand. The synthesis of
this strand requires many primers and, therefore, has DNA fragments with
intervening primers. These pieces are called Okazaki fragments because
the evidence supporting the discontinuous DNA synthesis was first provided by
Reiji and Tuneko Okazaki. The length of the fragments is 1000 to 2000
nucleotides.
The lagging strand has discontinuous
DNA due to the presence of primers. These primers are removed by the DNA polymerase-I
enzymes. This enzyme has the 5’ exonuclease and 3’ polymerase activity removing
nucleotides one by one from the 5’ end of the primer and adding nucleotides to
the 3’ end of the Okazaki fragment.
After the replacement of primers,
The DNA ligase enzyme joins the two DNA fragments by catalyzing the formation of the phosphodiester bond that seals
the nick between the discontinuously synthesized strands.
Proofreading and Error Correction
Although the action of DNA polymerases is accurate, sometimes synthesis is not perfect and a noncomplementary nucleotide is occasionally inserted. To remove these nucleotides the DNA polymerases possess 3′ to 5′ exonuclease activity. These enzymes detect and excise a mismatched nucleotide (in the 3′ to 5′ direction). Once the mismatched nucleotide is removed, 5′ to 3′ synthesis can again proceed.

No comments:
Post a Comment